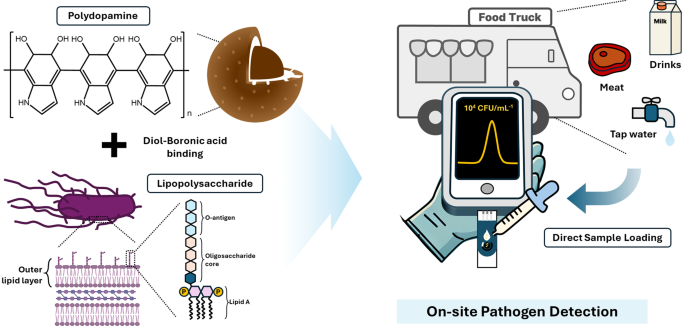
Determine 1 reveals a schematic of the extremely delicate and selective sensor for S. typhimurium, which was developed by imprinting LPS onto a PDA floor. Polydopamine is a wonderful template for MIP synthesis due to its versatile practical teams (catechol and amine), which allow sturdy interactions, tunable thickness relying on pH, and robust adhesion to varied surfaces [22]. Specifically, the thickness of the PDA coatings will be simply tuned primarily based on the pH situations throughout deposition [23]. This enables for exact management of the template layer thickness and optimization of the cavity construction within the ensuing MIP. Nanospherical MIPs with massive floor areas have been used to extend the variety of binding websites [24]. Sodium cyanoborohydride was used to stabilize the binding of FPBA to the PDA core, thereby enabling the formation of boronic acid on the floor for cis-diol interplay with LPS [19, 25]. Salmonella is managed in meals vans and eating places to make sure meals security by hygiene coaching, secure ingredient sourcing, correct cooking and storage temperatures, prevention of cross-contamination, common hand washing, thorough sanitation, and routine inspection [26]. Moreover, onsite sensor know-how for fast and straightforward Salmonella detection is important for enhancing meals security and stopping contamination.
Characterization of MIP
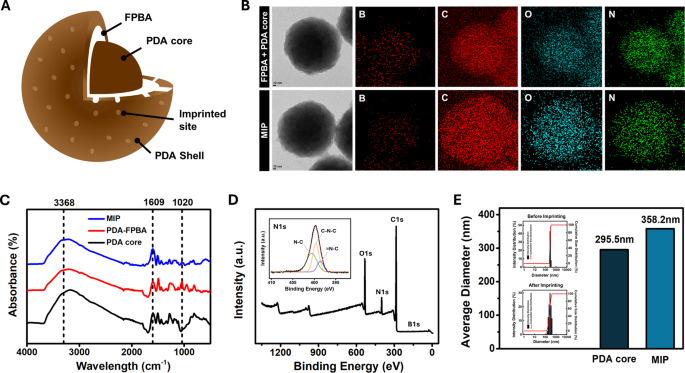
Determine 2A reveals the construction of the LPS-imprinted MIP. An LPS-recognizing MIP was produced by introducing 4-formylphenylboronic acid (FPBA) onto the floor of the PDA nanoparticles to bind with the cis-diol construction of LPS. A nanoscale PDA shell was subsequently shaped over the core particle and LPS was eluted to create binding websites on the MIP. Determine 2B reveals transmission electron microscopy (TEM) photos and energy-dispersive X-ray spectroscopy (EDS) mapping of the PDA core + FPBA and remaining MIP (PDA core + FPBA + PDA shell). After including FPBA to the PDA core, the atomic compositions of B, C, N, and O have been 10.3, 76.9, 4.1, and eight.8%, respectively. Following the formation of the PDA shell, the floor atomic compositions modified to eight.6%, 79.6%, 4.96%, and 6.79%, respectively. These modifications point out that the elevated boron atom composition confirmed the profitable formation of boronic acid on the PDA core floor, whereas the enrichment of C, N, and O validated the following formation of the PDA shell [22, 27]. As well as, the modifications within the practical teams all through the fabrication course of, from the PDA core to PDA-FPBA to the ultimate MIP, have been confirmed utilizing FT-IR (Fig. 2C). Attribute absorption bands of PDA within the PDA core and MIP have been recognized, together with 3368 cm⁻¹ for N-H stretching and 1609 cm⁻¹ for C = C stretching [28]. For PDA-FPBA, a peak equivalent to B-O-B stretching appeared at 1020 cm⁻¹, whereas the depth of the N-H and C = C stretching bands decreased, confirming the binding of FPBA to the PDA core [19]. XPS evaluation was carried out to look at the important parts current on the floor. The presence of C, O, N, and B within the PDA core, PDA-FPBA, and MIP was confirmed. The deconvoluted C 1s, O 1s, and N 1s spectra are proven in Figs. 2E and S2. Within the PDA-FPBA pattern, the lower in floor PDA in comparison with the core pattern was accompanied by a discount within the C = O, O = C, C = C, and C = N peaks. In step with the FT-IR outcomes, the modification of the PDA core with FPBA resulted within the look of a B 1s peak. Within the MIP pattern, the C = O, O = C, C = C, and C = N peaks have been partially recovered, most probably owing to the presence of the polydopamine shell [29, 30]. Due to this fact, the outcomes of the FTIR and XPS analyses supported the uniform incorporation of boronic acid and dopamine into the MIP.
For the imprinting time optimization, the thickness of shell ready with 2-hour and 1-hour, 45 min, and 30 min imprinting instances have been measured with DLS (Determine S3). It was decided that the MIP with 45 min imprinting time permits environment friendly LPS imprinting with out hindering subsequent LPS rebinding, exhibiting the most effective rebinding efficiency. As proven in Fig. 2D, the common MIP particle measurement elevated by roughly 50 nm from the PDA core measurement of 295.5 nm to 358.2 nm, suggesting that an imprinting time of 1 hour was optimum by the optimization course of.
MIP fabrication and characterization: (A) inner construction of MIP nanoparticle, (B) TEM photos and EDS mapping of FPBA + MIP core and MIP, (C) FT-IR spectra of MIP, PDA-FPBA, and PDA core, (D) XPS and high-resolution XPS spectra of N 1s area of MIP, and (E) DLS end result and histogram of PDA core and MIP
LPS detection efficiency of MIP sensor
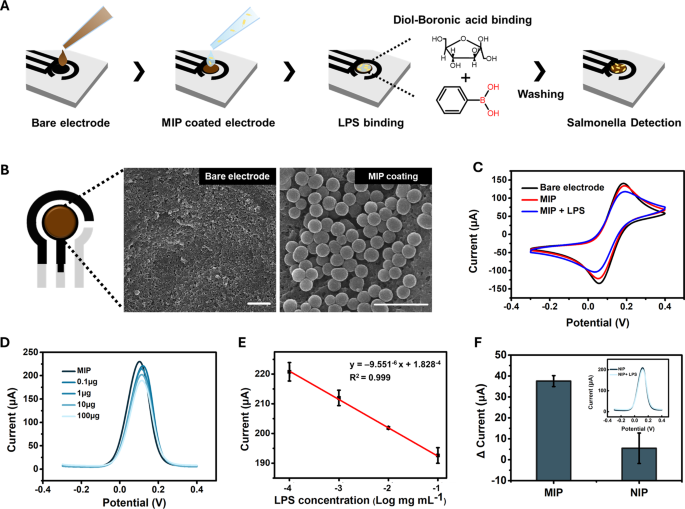
The sensing platform was fabricated utilizing a working electrode manufactured from single-walled carbon nanotubes (CNTs) and a counter electrode manufactured from carbon each of which exhibit excessive electrical conductivity in varied electrolytes [31, 32]. Ag/AgCl was used because the reference electrode. The working electrode was additional modified with the ultimate MIP utilizing drop-casting to judge its seize efficiency (Fig. 3A). The SEM picture in Fig. 3B reveals that the nanosphere-shaped MIP was uniformly connected to the CNT electrode.
To judge the sensing performance of Salmonella, the MIP-coated electrode was incubated with the LPS of S. typhimurium in PBS for 20 min, adopted by washing with PBS to take away the unbound LPS, as proven in Fig. 3A. The incubation time was optimized by measuring DPV at 10-minute intervals, and pH 7.4 PBS was used to forestall structural modifications in LPS (Determine S1). The binding of Salmonella LPS to SL-MIP was confirmed by cyclic voltammetry by evaluating the naked electrode (CTL), MIP-coated electrode (SL-MIP), and LPS-incubated MIP electrode (LPS + SL-MIP). As proven in Fig. 3C, the oxidation and discount peaks have been noticed at 0.185 and 0.057 V, respectively. Following MIP modification, a slight lower in peak present and a rise in ΔEpa have been noticed, indicating a minor improve in resistance attributable to the PDA layer. Notably, the addition of S. typhimurium LPS (1 mg/mL) to the SL-MIP electrode significantly diminished the oxidation and discount present peaks, indicating elevated resistance owing to LPS binding to SL-MIP.
Moreover, we carried out differential pulse voltammetry (DPV), which has increased sensitivity and discrimination of analytes than comparable oxidation potentials [33]. As proven in Fig. 3D, the DPV peak present at potentials of 0.11–0.12 V decreases with growing LPS focus, indicating the binding of LPS to the SL-MIP electrode. This concentration-dependent response exhibited a exceptional linear relationship between the logarithmic focus of LPS (0.1–100 µg) and the DPV peak present, with an R² worth exceeding 0.999 (Fig. 3E). The linear regression outcomes for Fig. 3E confirmed a p-value of 1.39(:instances:)10−7, indicating a extremely important correlation. When evaluating the responses of NIP and MIP to 100 µg of LPS, MIP exhibited a present change of 37.6 µA, whereas NIP exhibited a considerably decrease and inconsistent response of 5.5 µA (Fig. 3F). The p-value for the distinction between MIP and NIP was 0.043, confirming a statistically important distinction (p < 0.05). These findings demonstrated that the imprinting cavities on the LPS-mediated MIP floor have been extremely particular for Salmonella LPS. To additional consider the reproducibility and repeatability of the ready MIP sensor, similar electrodes have been fabricated with MIPs produced on completely different days and used to detect LPS at a 1.0 mg/mL focus (Determine S4). The height present values remained constant throughout the completely different electrodes, yielding a relative normal deviation (RSD) of 1.36%, demonstrating excessive reproducibility. In the meantime, the DPV measured from MIPs produced on the identical day exhibited minimal variation, a RSD of 1.98%, confirming repeatability.
LPS-detection efficiency of the SL-MIP: (A) experimental process for LPS and pathogen detection, (B) SEM picture of naked electrode and SL-MIP, scale bar(:=1:mu:m), (C) CV curves of naked electrode, MIP, and LPS-bound MIP at 50mVs−1, (D) DPV curves for the completely different concentrations of LPS from 0.1 to 100 (:mu:)g, (E) linear calibration curve of the SL-MIP for various LPS concentrations, and (F) present modifications of MIP and NIP for 100 (:mu:)g LPS
Salmonella detection efficiency of MIP sensor
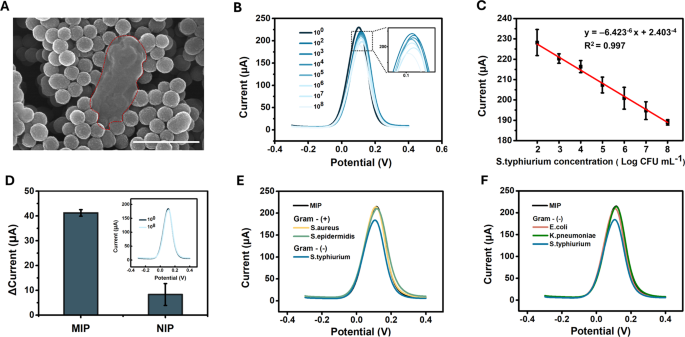
Figures 4A and S5 present an SEM picture of MIP incubated with S. typhimurium for 30 min, adopted by washing with PBS. In comparison with LPS, the floor of the entire cell displays a extra complicated construction, which can clarify the necessity for an extended incubation time to make sure enough binding energy (Determine S1). The areas marked with pink dashed traces point out the places of the bacterial cells or their remnants surrounded by MIP. When evaluating the quantitative detection functionality for entire S. typhimurium micro organism, growing cell focus resulted in a gradual lower in peak present at a possible of 0.11–0.12 V, just like the response of MIP to LPS (Fig. 4B). The coefficient of dedication (R2) was 0.997, indicating a powerful linear correlation between the logarithmic focus of S. typhimurium and the height present inside the 102–108 CFU/mL vary, with a detection restrict of 10 CFU/mL (Figs. 4C and S6). The linear regression outcomes for Fig. 4C confirmed a p-value of 4.42 (:instances:) 10−4, confirming a statistically important correlation. For the NIP, no important distinction was noticed between the present responses to PBS and S. typhimurium at 10⁸ CFU/mL (Fig. 4D). The p-value for the distinction between MIP and NIP was 0.0077, indicating a extremely important distinction (p < 0.01). To judge the selectivity, the response of the MIP sensor was examined towards varied bacterial strains generally present in meals samples. First, we in contrast S. typhimurium (a gram-negative bacterium) with S. aureus and S. epidermidis (gram-positive micro organism), which lack LPS on their cell surfaces. As proven in Fig. 4E, the MIP template generated by the S. typhimurium LPS exhibited no response to S. aureus or S. epidermidis (10⁸ CFU/mL). We investigated whether or not the MIP sensor may discriminate between gram-negative micro organism, contemplating the presence of the LPS on the cell floor. Notably, the MIP sensor responded solely to S. typhimurium and to not different gram-negative micro organism, similar to E. coli and Okay. pneumonia (Fig. 4F). This selectivity will be attributed to the species-specific variations within the O-polysaccharide chain, in addition to structural variations within the core and lipid A areas, which affect binding interactions with the imprinted cavities. Due to this fact, these findings recommend that the imprinted websites of MIP are extremely particular to S. typhimurium, highlighting the flexibility of the sensor to distinguish between LPS from varied bacterial species. Moreover, the reproducibility and repeatability of the sensor have been additionally evaluated for S. typhimurium at 107 CFU/mL (Determine S7). The height present measured utilizing MIPs produced on the identical day demonstrated a repeatability RSD of three.31%. When MIPs produced on completely different days have been examined, the reproducibility RSD was 2.14%, indicating excessive consistency throughout completely different manufacturing batches. Moreover, Cohen’s kappa evaluation was carried out to check the developed sensor with the normal tradition methodology [34]. The Cohen’s kappa index at 10² CFU/mL was 0.57, indicating reasonable settlement, which improved to substantial (103 CFU/mL) or nearly excellent (104 CFU/mL) settlement at increased inoculum concentrations, demonstrating the sensor’s excessive correlation with typical detection strategies.
Detection efficiency of the SL-MIP for S. typhimurium: (A) SEM picture of S. typhimurium cell traces surrounded by MIP nanoparticles, (B) DPV curves for various concentrations of S. typhimurium (from 100 to 108 CFU/mL), (C) linear calibration curve of the SL-MIP for various concentrations of S. typhimurium, (D) present modifications of MIP and NIP for 108 CFU mL− 1S. typhimurium, (E) DPV curves for gram-positive micro organism (S. aureus and S. epidermidis) and S. typhimurium at 108 CFU mL− 1, (F) DPV curves for gram-negative micro organism (E. coli and Okay. pneumoniae) and S. typhimurium at 108 CFU mL− 1
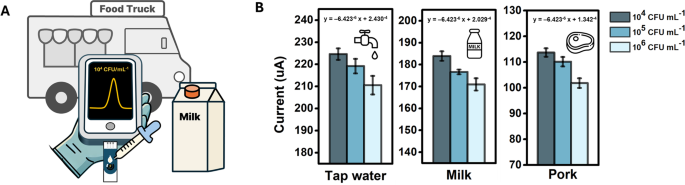
Salmonella detection efficiency of MIP sensor in meals matrices
Salmonella is managed in meals vans and eating places to make sure meals security by hygiene coaching, secure ingredient sourcing, correct cooking and storage temperatures, prevention of cross-contamination, common hand washing, thorough sanitation, and routine inspection [1]. Due to this fact, we investigated whether or not our MIP sensor may quickly detect S. typhimurium in actual meals samples for the on-site detection of foodborne pathogens. Determine S8 reveals the DPV response to varied concentrations of S. typhimurium in meals samples, together with faucet water, milk, and pork, whereas Fig. 5B illustrates the height present of the DPV corresponding to those concentrations [35, 36]. The concentrations have been chosen primarily based on ranges identified to trigger ailments in people. Notably, the MIP sensor exhibited a concentration-dependent response to the samples, even with out pre-treatment. A baseline shift was noticed owing to interfering with molecules in every pattern, which may alter the conductivity of the sensor. However, the diploma of the lower within the response present remained constant throughout S. typhimurium concentrations. Linear regression curves with similar slopes have been obtained for all of the meals samples, with corresponding R² values of 0.80, 0.90, and 0.87. To spotlight the efficiency of the ready MIP sensor, a comparability of the outcomes with not too long ago reported S. typhimurium sensors inside the final three years is offered in Desk 1.